Doing Bayesian Data Analysis
Thursday, December 7, 2023
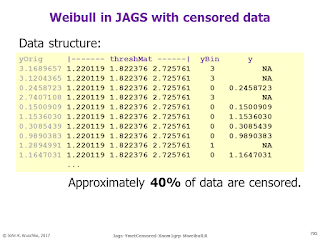
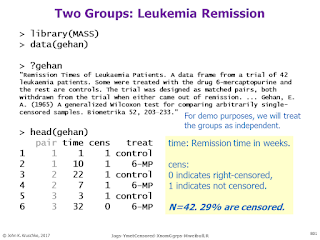
Bayesian survival analysis, using Weibull distribution and censored data
Sunday, November 26, 2023
From Stan forum: How to make decisions about results
Reposting from the Stan discussion site...
The original post:
Hi Everyone,
A question on how to handle reviewers when reporting CI. We are doing RL and in our papers when reporting results I always try to put a figure with the whole posterior. We also tend to report in text the median, 89%CI and probability of direction. We use 89% mainly as a way to avoid 95% which tend to encourage “black and white” thinking (89% is as arbitrary as any other value - but as McElreath said - its a prime number - so it’s easier to remember). A reviewer noted that we should change to 95% (see below). We worked really (really) hard on this piece and I don’t want to see this rejected. On the other hand - I won’t be able to look at myself if I just change to 95%. Obviously - no conclusion is going to change - its more a matter of principle.
Can you help me put forward good arguments?
Best,
NitzanReviewer:
There is only one thing that I would suggest the authors change, which is the use of 89% highest posterior density intervals. The authors cite McElreath’s “Statistical Rethinking”, which essentially states that 95% is arbitrary so we should be free to report any interval we see fit, and 89% has various properties that make it a satisfactory value. However, I would argue that the very fact that 95% is used near-universally (albeit originally quite arbitrarily) makes it the best value to use, as 1) it allows for easy comparison across studies, and 2) it is what people are used to and so facilitates straightforward and intuitive judgements about effects. It is also quite easy to miss that the CIs are 89% CIs and assume that they would be 95% CIs, and hence believe that the effects are stronger than they are.
My reply:
The Bayesian Analysis Reporting Guidelines (BARG) are intended to be helpful in cases like this. They’re published open-access here: https://www.nature.com/articles/s41562-021-01177-7 (Disclosure: I wrote the article.)
The BARG provide guidance for reporting decisions (among all other components of an analysis). First, do you really need to make decisions, or is the decision a ritual? If you do need to state decisions, then the BARG provide advice either using a credible interval (CI) or using posterior model probability.
If using a CI, the BARG are agnostic about what size of interval (or type of interval, HDI vs ETI) to use. The BARG emphasize that CI limits computed from MCMC are very wobbly because the limits are (typically) in low-density regions of posterior distribution; therefore high effective-sample size (ESS) is required. I think to more deeply justify the size of a decision interval you’d have to go full-blown decision theory with specified costs and benefits; for initial thoughts about doing that see the Supplement (to a different open-access article) available here: https://osf.io/fchdr. Meanwhile, in practice, I’d say to go with whatever size of interval you think is most useful for your research and your audience; after all, if they’re not convinced by a weaker decision criterion (i.e., only 89% instead of 95%) then your research will have less impact. I agree with zhez67373 regarding one possible option to satisfy both: put the audience-preferred criterion in the article and put your preferred criterion is supplemental material.
If using a Bayesian hypothesis test via model comparison, the BARG recommend reporting posterior model probabilities (not only Bayes factors, BFs) and basing a decision on the posterior model probability exceeding a criterion. Notice you need to specify a decision criterion for the posterior model probability; should it be 95%? 89%? Something else? A key issue here is that the posterior model probabilities depend on the assumed prior model probabilities. Therefore, the BARG recommend reporting the posterior model probabilities for a range of prior model probabilities, and, in particular, reporting the minimum prior probability the model could have and still meet the decision threshold. This concept is illustrated by Figure 2 in the BARG. (All of this assumes you have used appropriate prior distributions on parameters within the models; see the separate section in the BARG regarding prior sensitivity analysis.)
There is lots more discussion in the BARG.
P.S. I gave a talk about this at StanCon 2023.
P.P.S. I was not the reviewer!
Wednesday, August 30, 2023
New videos for Bayesian and frequentist side-by-side
There are new videos for Bayesian and frequentist side-by-side.
Eero Liski, a statistician at the Natural Resources Institute Finland, has made tutorial videos that introduce Bayesian and frequentist data analysis, side-by-side. The first of the video series is available here.
Thank you, Eero, for making these videos!
The videos were composed entirely independently from me, but they feature the R Shiny App I created a few years ago. The written (no-video) tutorial for that Shiny App is here.